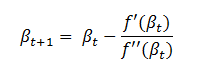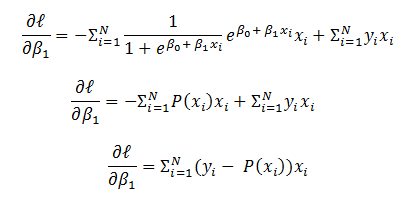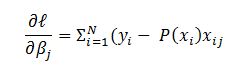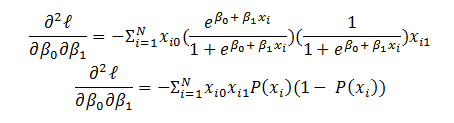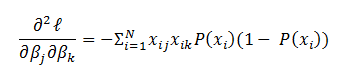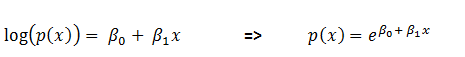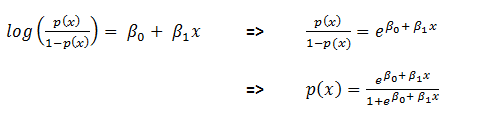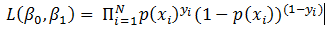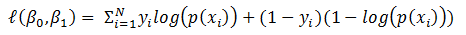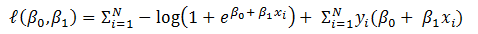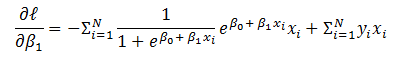Below are the objectives of this post:
- What is multi-layer feed-forward neural network
- Discuss back-propagation algorithm which is used to train it
- Implement what we discuss in python to gain better understanding
- Execute the implementation for a binary classification use-case to get a practical perspective
Multi-layer feed-forward neural network consists of multiple layers of artificial neurons. As the name suggests, one layer acts as input to the layer after it and hence feed-forward. But at the same time the learning of weights of each unit in hidden layer happens backwards and hence back-propagation learning.
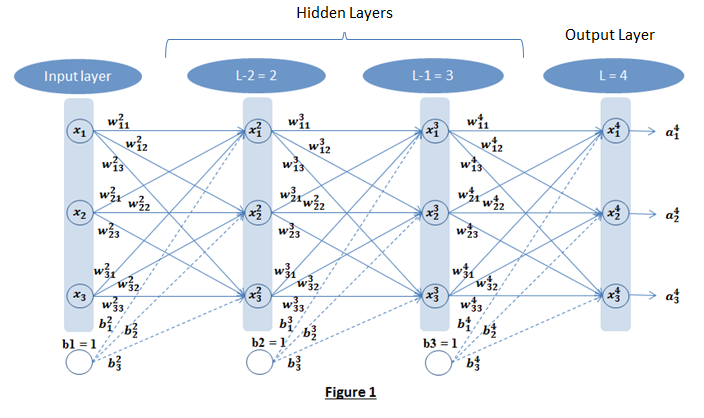
For this discussion let us consider the neural network shown in Figure 1. It has three neural units in each layers and b1, b2, b3 are included as units (with input as 1) for bias. Figure 1 has one input layer, one output layer (layer L) and 2 hidden layers (L-1 and L-2).
Below are two high level steps in building a multi-layer feed-forward neural network model. These steps are executed iteratively:
- Feed-forward: Data from input layer is fed forward through each layer and then output is generated in the final layer.
- Back-propagation: This is the learning step. Once the output is generated, the error is calculated w.r.t. the expected output and then this error is propagated in backwards direction to adjust the weights to reduce the error. This is the learning step.
Let us discuss each of the steps in detail:
- Feed-forward: On a high-level two steps are executed at every unit during feed-forward:
- Weighted sum of all inputs is calculated (including bias). Figure 2 shows a detailed view of
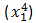 unit.
unit. 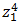 is the input to
is the input to 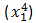 and is weighted sum of all three activations (non-linearized output) and the bias from the previous layer i.e.
and is weighted sum of all three activations (non-linearized output) and the bias from the previous layer i.e. 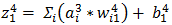
- Then non-linearization of the input (calculated in #1 above) is done i.e.
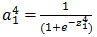 . Here
. Here 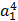 is the output of the unit as shown in Figure 2.
is the output of the unit as shown in Figure 2. 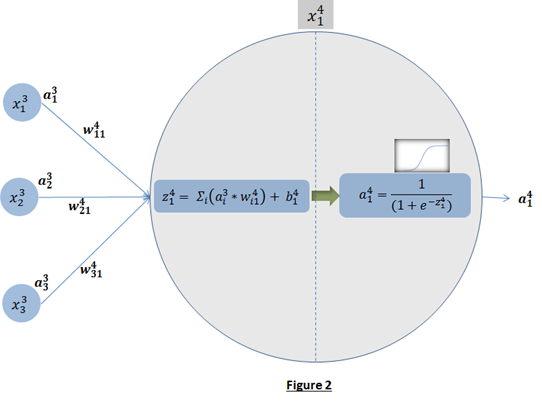 Similarly, each unit receives the inputs from previous layers and passes the non-linearized output to the next layer. The functions for non-linearization are chosen such that they are differentiable (we will answer ‘why?’ while talking about back-propagation). Below are few notations to keep in mind for generalization:
Similarly, each unit receives the inputs from previous layers and passes the non-linearized output to the next layer. The functions for non-linearization are chosen such that they are differentiable (we will answer ‘why?’ while talking about back-propagation). Below are few notations to keep in mind for generalization:
- Weighted sum of all inputs is calculated (including bias). Figure 2 shows a detailed view of
- Back-propagation learning: The overall objective of backpropagation learning is to learn combination of weights which minimizes the output error. The error between actual output and the desired output is reduced (in each iteration) by doing supervised adjustment in weights. We will not be able to visualize the space with all weights and error but for intuitive understanding let’s see error with two weights in Figure 3. Objective is to learn weights such that value of E is minimum (as pointed by arrow in Figure 3).
 Now the question is, how to learn this combination of weights? We will do this learning using gradient descent so that weights learnt, in each iteration, move ‘generated output’ in the direction towards the ‘expected output’. As gradient descent requires the cost function to be differentiable, we have taken sigmoid as activation function.Now let us write the above understanding in maths so that we can implement it later in python. For neural network in Figure 1, the error function is defined as:
Now the question is, how to learn this combination of weights? We will do this learning using gradient descent so that weights learnt, in each iteration, move ‘generated output’ in the direction towards the ‘expected output’. As gradient descent requires the cost function to be differentiable, we have taken sigmoid as activation function.Now let us write the above understanding in maths so that we can implement it later in python. For neural network in Figure 1, the error function is defined as: 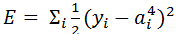 where
where 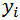 is the expected output of ith unit of output layer. We need to find
is the expected output of ith unit of output layer. We need to find 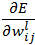 for all units of all layers and adjust each weight as below:
for all units of all layers and adjust each weight as below:
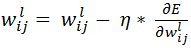
where 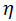 is the learning rate.
is the learning rate.
Let us derive an expression for 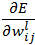 which can be generalized to any weight and any layer in the network.
which can be generalized to any weight and any layer in the network.
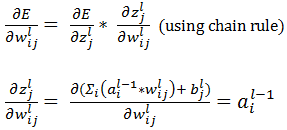
Let us call 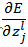 as
as 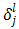 , which represents change in error with unit change in input to jth unit of lth layer. So,
, which represents change in error with unit change in input to jth unit of lth layer. So,

Let us compute 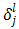 if l = ‘L’ i.e. the output layer for any neural network (L= 4 in Figure 1) then,
if l = ‘L’ i.e. the output layer for any neural network (L= 4 in Figure 1) then,
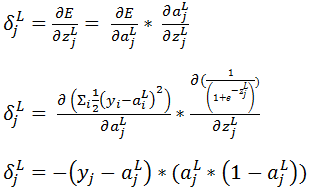
Now let’s derive an expression for 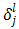 for hidden layers:
for hidden layers:
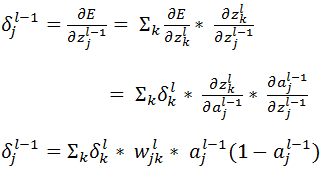
Based on the above equation we can calculate 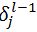 for all units of all layers, provided the same has been calculated for the layer ahead. So we will have to calculate it backwards. So let’s conclude:
for all units of all layers, provided the same has been calculated for the layer ahead. So we will have to calculate it backwards. So let’s conclude:
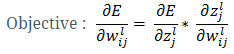
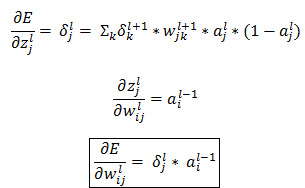
Similarly, we will derive expression for 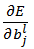 as:
as:
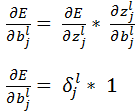

Finally, the weight updates happens as earlier discussed:
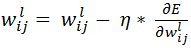
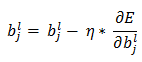
For online learning, one example is introduced at a time and weights are updated incrementally (for each example).
For batch learning, a batch of examples is introduced and weights are updated only once for the whole batch. So an average of gradients for all examples is used to update the weights.
Below is the python implementation for feed-forward backpropagation in batch learning mode:
"""
Author: Ashish Verma
This code was developed to give a clear understanding of what goes behind the curtains in multi-layer feedforward backpropagation neural network.
Feel free to use/modify/improve/etc.
Caution: This may not be an efficient code for production related usage so thoroughly review and test the code before any usage.
"""
import numpy as np
import matplotlib.pyplot as plt
from sklearn import preprocessing
from scipy import spatial
class multiLayerFeedForwardNN:
def __init__(self, listOfNeuronsEachLayer):
self.listOfNeuronsEachLayer = listOfNeuronsEachLayer
self.dictOfWeightsEachLayer = {}
self.dictOfActivationsEachLayer = {}
self.dictOfDeltaEachLayer = {}
self.dictOfZeeEachLayer = {}
self.dictOfGradientsEachLayer = {}
self.listAvgCost = []
#Initialize weights with random values (this also includes bias terms)
for iLayer in range(1,len(listOfNeuronsEachLayer)):
self.dictOfWeightsEachLayer[iLayer + 1] = np.mat(np.random.random((listOfNeuronsEachLayer[iLayer], listOfNeuronsEachLayer[iLayer-1]+1)))
def sigmoidFunc(self, z):
result = 1/(1 + np.exp(-1*z))
return result
def feedForward(self, inputChunk):
#Loop over all layers
for iLayer in range(1, len(self.listOfNeuronsEachLayer)):
#Save input itself as the activation for the input layer. We will use it later in the code
if(iLayer == 1):
self.dictOfActivationsEachLayer[1] = inputChunk
#Insert extra element for bias having value 1 in input elements
inputChunk = np.hstack([inputChunk, np.mat(np.ones(len(inputChunk))).T])
#Calculate z using matrix multiplication of weights and inputs
self.dictOfZeeEachLayer[iLayer+1] = np.dot(inputChunk, self.dictOfWeightsEachLayer[iLayer+1].T)
#Calculate 'a' after non-linearizing z
self.dictOfActivationsEachLayer[iLayer+1] = self.sigmoidFunc(self.dictOfZeeEachLayer[iLayer+1])
#Consider activation of the current layer as input to the next layer
inputChunk = self.dictOfActivationsEachLayer[iLayer+1]
def calculateDelta(self, outputChunk):
#Calculate delta backwards (output layer to 2nd layer)
for iLayer in range(len(self.listOfNeuronsEachLayer),1,-1):
#For last layer calculate delta
if(iLayer == len(self.listOfNeuronsEachLayer)):
self.dictOfDeltaEachLayer[iLayer] = np.multiply((self.dictOfActivationsEachLayer[iLayer] - outputChunk), np.multiply(self.dictOfActivationsEachLayer[iLayer],(1-self.dictOfActivationsEachLayer[iLayer])))
#For rest of the layers calculate delta using delta of next layer
else:
wDelta = np.dot(self.dictOfDeltaEachLayer[iLayer+1], np.delete(self.dictOfWeightsEachLayer[iLayer + 1],self.dictOfWeightsEachLayer[iLayer + 1].shape[1]-1,1))
dadz = np.multiply(self.dictOfActivationsEachLayer[iLayer],(1-self.dictOfActivationsEachLayer[iLayer]))
self.dictOfDeltaEachLayer[iLayer] = np.multiply(wDelta, dadz)
def calculateErrorGradient(self):
#Calculate error gradient for all layers
for iLayer in range(2,len(self.listOfNeuronsEachLayer)+1):
#Gradient w.r.t all weights
self.dictOfGradientsEachLayer[iLayer] = np.dot(self.dictOfDeltaEachLayer[iLayer].T,self.dictOfActivationsEachLayer[iLayer-1])
#Gradient w.r.t. biases and add it to the other gradients
self.dictOfGradientsEachLayer[iLayer] = np.hstack([self.dictOfGradientsEachLayer[iLayer], np.sum(self.dictOfDeltaEachLayer[iLayer],0).T])
def updateWeights(self, learningRate):
#For all layers update weights using average of gradients over all examples
for iLayer in range(2, len(self.listOfNeuronsEachLayer)+1):
self.dictOfWeightsEachLayer[iLayer] = self.dictOfWeightsEachLayer[iLayer] - (learningRate/len(self.dictOfActivationsEachLayer[1]))*self.dictOfGradientsEachLayer[iLayer]
def calculateAverageCost(self, actualOutput, expectedOutput):
#Calculate average of error function
avgOverLayer = np.average([(1/2.)*x**2 for x in np.array(actualOutput-expectedOutput)],1)
return np.average(avgOverLayer)
def trainBatchFFBP(self, learningRate, inputData, outputData, miniBatchSize, maxIterations):
#If the size of the data is large and resources are less, then divide it into batches for computation.
#Define batch size accordingly or leave empty if data size is not big
if(miniBatchSize == ''):
miniBatchSize = len(inputData)
#Initialize iteration counter
iterations = 0
#Iterate over all examples 'maxIterations' number of times
#We can even let the program iterate untill the desired error reduction is achieved (this code doesn't implement it).
for iterations in range(maxIterations):
#iBatch takes care of dividing the full data in batches (if we chose to, incase the data is big)
startRecord = 0
for iBatch in range((len(inputData)/miniBatchSize)):
endRecord = startRecord + miniBatchSize
inputChunk = inputData[startRecord:endRecord]
outputChunk = outputData[startRecord:endRecord]
self.feedForward(inputChunk)
self.calculateDelta(outputChunk)
self.calculateErrorGradient()
self.updateWeights(learningRate)
startRecord = endRecord
#print "iBatch", iBatch
#--------------Just stack up all calculated output for average error calculation-----------------#
if(iBatch == 0):
activationMat = self.dictOfActivationsEachLayer[len(self.listOfNeuronsEachLayer)].copy()
else:
activationMat = np.vstack([activationMat,self.dictOfActivationsEachLayer[self.listOfNeuronsEachLayer]])
#------------------------------------------------------------------------------------------------#
if(endRecord < len(inputData)):
inputChunk = inputData[startRecord:]
outputChunk = outputData[startRecord:]
self.feedForward(inputChunk)
self.calculateDelta(outputChunk)
self.calculateErrorGradient()
self.updateWeights(learningRate)
activationMat = np.vstack([activationMat,self.dictOfActivationsEachLayer[self.listOfNeuronsEachLayer]])
self.listAvgCost.append(self.calculateAverageCost(activationMat, outputData))
iterations = iterations + 1
print iterations
Now, let us use the implementation for a binary classification task. We will be using the dataset obtained from https://archive.ics.uci.edu/ml/datasets/Ionosphere.
Ionosphere dataset’s output is either ‘g’ (good) or ‘b’ (bad). Let’s replace ‘g’ by 1 and ‘b’ by 0. This dataset has 34 features as input, so the input layer will have 34 neurons and we are using sigmoid activation function so we will be taking 1 neuron in output layer for binary classification.
#Import dataset
dataset = np.genfromtxt ('C:\Research\BlogPloration\BackPropogation\ComparisonDataset\ionosphere.csv', delimiter=",")
#Initialize 'multiLayerFeedForwardNN' with 34, 27 and 1 neurons in input, hidden and output layers respectively
objMLFFNN = multiLayerFeedForwardNN([34,27,1])
#Start training with learning rate as 0.5 and 120,000 iterations
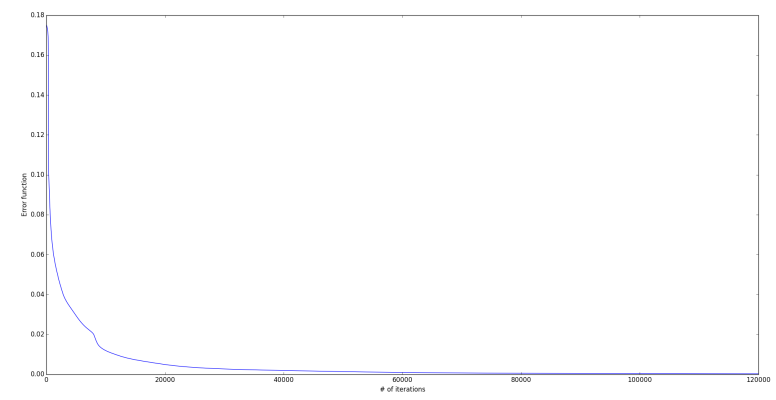
objMLFFNN.trainBatchFFBP(0.5, np.mat(dataset[:,:34]), np.mat(dataset[:,34]).T,'', 120000)
This NN model fits the data fully. Let’s look at the plot of ‘error function value’ calculated with iterations during training. Error for every iteration is stored in ‘listAvgCost’.
Note: In order to keep things simple, I haven’t talked about regularization term and other aspects. I would suggest the readers to study further once the basic is ‘very’ clear.